Positive strand RNA viruses as emerging human pathogens
COVID-19 pandemic has revealed a magnitude of problems that can be caused by single emerging viral pathogens. SARS-CoV-2 belongs to family Coronaviridae, one of the families of positive-strand RNA viruses. Viruses with positive-strand RNA genomes are, however, diverse and widespread in nature. Many of them are major human pathogens causing millions of cases each year: seasonal coronaviruses, dengue virus (DENV), noroviruses, rhinoviruses to name just few. Others have known to cause sudden outbreaks and epidemics - recent examples include Zika virus (ZIKV, family Flaviviridae, genus Flavivirus) outbreak 2015-2016 and chikungunya virus (CHIKV, family Togaviridae, genus Alphavirus) outbreaks 2005-2008 and 2013. List of positive-strand RNA viruses that have potential to cause such outbreaks/epidemics/pandemics is much longer and furthermore, many viruses with such a potential are currently poorly studied or simply unknown.
Viruses causing epidemics use different modes of transmission. These include droplet/aerosol transmission (SARS-CoV-2) and transmission by arthropod vectors (ZIKV, CHIKV). Historically, our research group has been engaged in basic and applied studies of alphaviruses that are pathogenic for humans: CHIKV, Ross River virus (RRV), Barmah Forest virus (BFV), o’nyong’nyong virus (ONNV), Eastern equine encephalitis virus (EEEV), Sindbis virus (SINV) and also Semliki Forest virus (SFV), not associated with known diseases. During the ZIKV outbreak, our research interest was extended to flaviviruses and, by obvious reasons, at 2020 extended again to include SARS-CoV-2. Previously we have also worked with hepatitis C virus (HCV) and human immunodeficiency virus type I (HIV-1). We maintain research competence with those viruses.
A general overview of our current areas of research and expertise can be found below with a selection of appropriate publications listed. All figures and illustrations are linked to their respective sources and can be accessed by clicking the images.
Functional studies of alphavirus replicase proteins
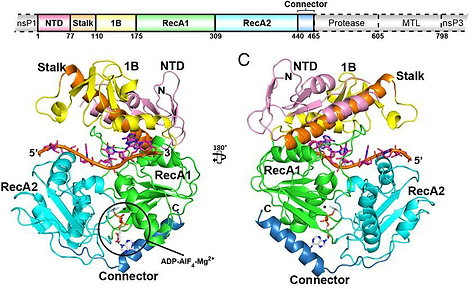
This has been the main topic for the RNA virus research group since its establishment at 2003; prof Andres Merits has been working with alphavirus replicase, its components, their properties and interactions even before that time. It is also area of key competences and expertise for the research group. The main research approaches have been development of reverse genetics for these viruses and tools for in depth studies of the viral replicase. We have been working with all four replicase proteins (nsP1, nsP2, nsP3 and nsP4), have developed methods of purification and enzymatic assays for all of them and participated in studies resulting in revealing 3D-structures of these enzymes. The main focus has been nsP2, the viral RNA helicase/protease and also the key enzyme involved in regulation of alphavirus replication.
-
Das, P.K., Merits, A. & Lulla A. (2014). Functional crosstalk between distant domains of chikungunya virus non-structural protein 2 Is decisive for its RNA-modulating activity. Journal of Biological Chemistry. 289(9):5635-5653. PMCID: PMC3937639
-
Abraham, R., Hauer, D., McPherson R.L., Utt, A., Kirby, I., Cohen, M.S., Merits, A, Leung, A.K.L.& Griffin, D.E. (2018) ADP ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proceedings of the National Academy of Sciences of the USA, 115(44):E10457-E10466. PMCID: PMC6217424
-
Bakhache, W., Neyret, A., Bernard, E., Merits, A. & Briant, L. (2020). Palmitoylated cysteines in chikungunya virus nsP1 are critical for targeting to cholesterol-rich plasma membrane microdomains with functional consequences for viral genome replication. Journal of Virology, 94(10):e02183-19. PMCID: PMC7199415
-
Law, Y.-S., Utt, A., Tan, Y.B., Zheng, J., Wang, S. Chen, M.W., Griffin, P.R., Merits, A. & Luo, D. (2019). Structural insights into RNA recognition by the chikungunya virus nsP2 helicase. Proceedings of the National Academy of Sciences of the U S A, 116(19):9558-9567. PMCID: PMC6511008
-
Tan, Y.B., Lello, L.S., Liu, X., Law, Y.-S., Kang, C., Lescar, J., Zheng, J., Merits, A. & Luo, D. (2022). Crystal structures of alphavirus nonstructural protein 4 (nsP4) reveal an intrinsically dynamic RNA-dependent RNA polymerase fold. doi: 10.1093/nar/gkab1302. PMCID: PMC8789068
Despite of decades work in this area, our knowledge in this area is still far from being complete. Current research topics include structure-guided analysis of critical determinants in alphavirus replicase proteins and development of cell-based assays to screen for chemical compounds inhibiting virus-encoded enzymes. We have demonstrated that virus genomes containing mutations affecting activities of these enzymes are often attenuated; therefore we are also involved in studies aiming the use of these attenuated viruses as anti-alphavirus vaccine candidates.
Assembly of functional alphavirus replicase complex
In order to form the functional replicase complex alphavirus replicase proteins should interact with each other and with the virus genome. RNA virus lab has been involved in studies that have revealed that in order to do so the proteins need to be produced in form of polyprotein precursors that are processed by nsP2 protease activity in a highly specific manner, and that disturbance of this processing pathway has major consequences for virus infectivity and virulence.
-
Hellström, K., Kallio, K., Utt, A., Quirin, T., Jokitalo, E., Merits, A. & Ahola, T. (2017). Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. Journal of Virology, 91(18):e00787-17. PMCID: PMC5571266
-
Lulla, V., Karo-Astover, L., Rausalu, K., Saul, S., Merits, A. & Lulla, A. (2018). Timeliness of proteolytic events is prerequisite for efficient functioning of the alphaviral replicase. Journal of Virology, 92(14):e00151-18. PMCID: PMC6026757..
-
Lello, L.S., Utt, A., Bartholomeeusen, K., Wang, S., Rausalu, K., Kendall, C., Coppens, S., Fragkoudis, R., Tuplin, A., Alphey, L., Ariën, K.K. & Merits, A. (2020). PMC7498090
-
Teppor, M., Žusinaite, E., Karo-Astover, L., Omler, A., Rausalu, K., Lulla, V., Lulla, A. & Merits, A. (2021). Semliki Forest virus chimeras with functional replicase modules from related alphaviruses survive by adaptive mutations in functionally important hotspots. Journal of Virology, doi: 10.1128/JVI.00973-21
Current research topics include analysis of the molecular basis of compatibility of alphavirus replicase complex components. We are also working with viral genomic RNA which is also a crucial component of replicase complex and regulates its formation. This work includes mapping of determinants responsible for alphavirus RNA template recognition, gene expression and RNA replication as well as analysis of functional roles of RNA secondary structures, long-range interactions and interactions with viral and host components. In addition to these basic studies, we are applying obtained knowledge and experience for development of advanced tools for detection of alphavirus infection. These systems can also be used as tools to limit virus transmission. Similarly, the role of nsP2 in regulation of virus replicase formation can be turned against the virus and used for development of genetically modified mosquitoes that are unable to transmit alphaviruses.
Elucidating host-virus interactions that contribute to alphavirus replication and pathogenesis
Our studies of the functions of replicase proteins and virus replication complexes lead to the analysis of the complicated interactions between alphavirus and the host. We have studied effects of mutations in alphavirus replicase proteins on the ability of virus to persist in infected host cells, and we have identified numerous host protein interactions with alphavirus replicases. Some of these interactions are essential for the virus and some (such as activation of interferon response) are generally regarded as harmful for the virus. However, our studies have clearly shown that this is not strictly true, some interactions (such as activation of interferon responses) can also represent a part of virus attack strategy and be related to viral pathogenesis.
-
Nikonov, A., Mölder, T., Sikut, R., Kiiver, K., Männik, A., Toots, U., Lulla, A., Lulla, V., Utt, A., Merits, A. & Ustav, M. (2013). RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. , 9, e1003610. doi: 10.1371/journal.ppat.1003610.
-
Maillard, P.V., Van der Veen, A.G., Deddouche-Grass, S., Rogers, N.C., Merits, A. & Reis E Sousa, C. (2016). Inactivation of the type I interferon pathway reveals long dsRNA-mediated RNA interference. The EMBO Journal, 35(23):2505-2518. PMCID: PMC5167344
-
Liu, X., Mutso, M., Utt. A., Lepland, A., Herrero, L., Taylor, A., Bettadapura, J., Rudd, P., Merits, A. & Mahalingam, S. (2018). Decreased virulence of Ross River virus harboring mutation in the first cleavage site of non-structural polyprotein is caused by a novel mechanism leading to increased production of interferon-inducing RNAs. , 9(4):e00044-18. PMC6106088.
-
Götte, B., Utt, A., Fragkoudis, R., Merits, A. & McInerney, G. (2020). Sensitivity of alphaviruses to G3BP deletion correlates with efficiency of replicase polyprotein processing. Journal of Virology, 94(7):e01681-19. PMCID: PMC7081891.
-
Wang, S. & Merits, A. (2022). G3BP/Rin-binding motifs inserted into flexible regions of nsP2 support RNA replication of chikungunya virus. Journal of Virology, 96(21):e0127822. doi: 10.1128/jvi.01278-22, PMCID: PMC9645214
Hundreds of host proteins interact with alphavirus replicase complexes. However, the mechanisms of their action (and consequences of lack of these interactions) are generally not known. The detailed analysis of these mechanisms is complicated and, for arboviruses, there are differences between the interactions in cells of vertebrate host and arthropod vector. Our interest is currently focused on two factors interacting with the hypervariable domain of nsP3. First, G3BP proteins (Rasputin/Rin in mosquitoes) are absolutely required for CHIKV replication but dispensable for some other alphaviruses. The G3BPs do not affect the switch from translation to RNA replication or the processing of CHIKV replicase precursor; however, in their absence CHIKV is unable to synthesize negative-strand RNA. We hypothesize that G3BPs may be involved in the binding of viral replicase to genomic RNA and/or regulate the timing of different steps essential for replicase complex formation. FHL1 (and related proteins) are also involved, albeit not absolutely essential, in CHIKV replication. We participate in studies aiming analysis of the impact of FHL1 for replication of CHIKV in vitro and in vivo. The CHIKV mutants harboring mutation affecting ability of nsP3 to interact with G3BPs or FHL1 are evaluated for potential to be used as anti-alphavirus vaccines.
Analysis of the mechanism used by host and transmission vectors to limit alphavirus infection and pathology
The increasing importance of alphaviruses, most notably CHIKV, promoted novel directions including analysis of host mechanisms counter-acting alphavirus infection. Our research group has (mostly in international collaborations) worked on characterization of host defense mechanisms against alphaviruses and counter-mechanisms used by alphaviruses. These studies have also revealed the complicated roles of host immunity that, as for many viruses, also impacts alphavirus-induced pathology.
-
Teng, T.S., Foo, S.S., Simamarta, D., Lum, F.M., Teo, T.H., Lulla, A., Yeo, N.K., Koh, E.G., Chow, A., Leo, Y.S., , Chin, K.C. & Ng, L.F.P. (2012). Viperin restricts chikungunya virus replication and pathology. Journal of Clinical Investigations, 122(12): 4447-60. PMCID: PMC3533538
-
Pingen, M., Bryden, S.R., Pondeville, E., Schnettler, E., Kohl, A., Fazakerley, J.K., Graham, G.J. & McKimmie, C.S. (2016). Host inflammatory response to mosquito bites defines severity of arbovirus infection. 44(6):1455-69. PMC4920956
-
Bryden, S.R., Pingen, M., Lefteri, D.A., Major, J., Delang, L., Jacobs, S., Abdelnabi, R., Neyts, J., Miltenburg, J., Khalid, H., Tuplin, A., Merits, A., Pondeville, E., Edgar, J., Graham, G.J., Shams, K. & McKimmie, C.S. (2020). Pan-viral protection against arboviruses by targeting inoculation site-based skin macrophages. Science Translational Medicine, 12(527):eaax2421. doi: 10.1126/scitranslmed.aax2421
-
Lefteri, D.A., Bryden, S.R., Pingen, M., Terrry, S., McCafferty, A., Beswick, E.,
Georgiev, G., Van Der Laan, M., Mastrullo, V., Campagnolo, P., Waterhouse, R.M., Varjak, M., Merits, A., Fragkoudis, R., Griffin, S., Shams, K., Pondeville, E. & McKimmie, C.S. (2022). Mosquito saliva enhances virus infection through sialokinin-dependent vascular leakage. Proceedings of the National Academy of Sciences of the USA, 119(24):e2114309119. doi: 10.1073/pnas.2114309119. PMCID: PMC9214539 -
Jungfleisch, J., Böttcher, R., Talló-Parra, M., Pérez-Vilaro, G., Merits, A., Novoa, E.M. & Diez Anton, J. (2022). CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. 13(1):4725.
We are working on molecular bases of alphavirus vector transmission on examples of CHIKV and ONNV. CHIKV is transmitted by Aedes mosquitoes while ONNV uses mostly anthropophilic Anopheles gambiae. We have found evidence that RNA replication plays a crucial role in vector specificity of CHIKV and ONNV. In part, this is due to differences in nsP3 proteins of these viruses and may be (or not) connected with use of Rasputin/Rin proteins from different mosquito species. These topics are studied using chimeric viruses and their replicases in Aedes and Anopheles cells. Another topic active in the lab is development of sensors for virus infection. When used in transgenic mosquitos such sensors should detect activities of viral proteins (for example viral proteases) or replication complexes and respond with expression of proteins killing infected cell (or mosquito). We consider this a promising approach to limit ability of mosquitoes to transmit pathogenic viruses.
Development of tools and assays applicable for characterization of mechanisms of action of inhibitors of alphavirus RNA replicase
The increasing importance of alphaviruses (particularly CHIKV) also promoted novel directions aimed to counter-act CHIKV infection and to develop antiviral strategies such as antiviral compounds. Advantage was taken from our basic studies on the structure and function of alphavirus replicase proteins. In addition, based on our insights into the pathways of virus replicase complex formation we have developed multiple tools and assays applicable to the screening of antiviral compounds, analysis of their mechanism of action, and characterization of resistance against these compounds.
-
Pohjala, L., Utt, A., Varjak, M., Lulla, A., Merits, A., Ahola, T. & Tammela, P. (2011). Inhibitors of alphavirus entry and replication identified with a stable chikungunya replicon cell line and virus-based assays. , 6(12):e28923. PMCID: PMC3242765
-
Das, P.K., Puusepp, L., Varghese, F.S., Utt, A., Ahola, T., Kananovich, D.G., Lopp, M., Merits, A. & Karelson, M. (2016). Design and validation of novel chikungunya virus protease inhibitors. , 60(12):7382-7395. PMC5119020.
-
Teo, T.H., Chan, Y.H., Lee, W.W., Lum, F.M., Amrun, S.N., Her, Z., Rajarethinam, R., Merits, A., Rötzschke, O., Rénia, L. & Ng, L.F.P.(2017). Fingolimod treatment abrogates chikungunya virus-induced arthralgia. Science Translational Medicine, 9(375):eaal1333. doi: 10.1126/scitranslmed.aal1333
-
Varghese, F.S., Rausalu, K., Hakanen, M., Saul, S., Kümmerer, B., Susi, P., Merits, A. & Ahola, T. (2017). Obatoclax inhibits alphavirus membrane fusion by neutralizing the acidic environment of endocytic compartments. , 61(3):e02227-16. PMC5328557
-
Varghese, F.S., Meutiawati, F., Teppor, M., Jacobs, S., de Keyzer, C., Taskoprü, E., van Woudenbergh, E., Overheul, G.J., Bouma, E., Smit, J.M., Delang, L., Merits, A. & van Rij, R.P. (2022). Posaconazole inhibits multiple steps of the alphavirus replication cycle. 197:105223.
Screening for antiviral compounds using infectious viruses tends to identify large number of compounds that affect virus indirectly by altering cellular processes used by a virus. Though such compounds, also called host-targeting antivirals, have broad-spectrum antiviral activities they suffer from low selective indexes. As the compounds target a host cell their specificity is typically hard to increase. Hits identified using replicon-based screening are more often directly acting antivirals; however, typically their targets are not known. Identification of targets of these hit compounds as well as conformation of antiviral activity of rationally designed inhibitory compounds represent important steps for antiviral drug development. We are working on development/optimization of test-tube based assays designed to confirm targeting of activities of nsP1, nsP2 and nsP4 and cell-based assays to do the same. In addition, we are developing new virus-based tools (recombinant viruses with marker genes) that allow conformation of antiviral effect of compounds in virus infected cells or animals.
Flavivirus infection and transmission
Outbreak of ZIKV has boosted our interest to this pathogen and flaviviruses in general. In a very short time we have been able to contribute to the filed by developing a new ZIKV reverse genetics system (currently most commonly used system in world, at least 50 different laboratories have requested and obtained it form us), applying it for studies of anti-ZIKV compounds; developing of novel assay for diagnostics of ZIKV infection and by analyzing ZIKV/mosquito vector interactions. Subsequently we extended our studies to different mosquito- and tick-born flaviviruses including dengue virus, West Nile virus, yellow fever virus, duck tembusu virus, tick-born encephalitis virus and Langat virus.
-
Mutso, M., Saul, S., Rausalu, K., Susova, O., Žusinaite, E., Mahalingam, S. & Merits, A. (2017). Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on Brazilian Zika virus isolate. , 98, 2712-2724. doi: 10.1099/jgv.0.000938.
-
Lum, F.-M., Lin, C., Susova, O.Y, Teo, T.-H., Fong, S.-W., Mak, T.-M., Lee, L.K., Chong, C.-Y., Lye, D.C.B., Lin, R.T.P., Merits, A., Leo, Y.-S. and Ng, L.F.P. (2017). Sensitive detection of Zika virus antigen in patients’ whole blood as an alternative diagnostic approach. , 216, 182-190. doi: 10.1093/infdis/jix276
-
Varjak, M., Donald, C.L., Mottram, T., Sreenu, V.B., Merits, A., Maringer, K., Schnettler, E. and Kohl, A. (2017). Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. , 11:e0006010. doi: 10.1371/journal.pntd.000601
-
Mutso M., St John, J.A., Ling, Z.L., Burt, F.J., Poo, Y.S., Liu, X., Žusinaite, E., Grau, G.E., Hueston, L., Merits, A., King, N.J.C., Ekberg, J.A.K. and Mahalingam, S. (2020) Basic insights into Zika virus infection of neuroglial and brain endothelial cells. Journal of General Virology. 101(6)622-634. doi: 10.1099/jgv.0.001416.
-
Gestuveo, R.J., Royle, J., Donald, C.L., Lamont, D.J., Hutchinson, E.C., , Kohl, A. and Varjak, M. (2021). Analysis of Zika virus capsid-Aedes aegypti mosquito interactome reveals pro-viral host factors critical for establishing infection. Nature Communications, 12(1):2766. doi: 10.1038/s41467-021-22966-8.
Several projects concerning flaviviruses are currently ongoing. First, we continue development of reverse genetic systems for flaviviruses of interest. To make their studies more efficient we are developing approaches insertion of sequences encoding for markers (reporter proteins or epitope tags) into flavivirus genomes and perform analysis of viability and genetic stability of obtained recombinant viruses. Such viruses can be used for analysis of virus infection as well as for screening of antiviral compounds. Second, we are developing cell lines harboring flavivirus replicons (genomes lacking part of viral glycoproteins) that can be used both for studies of flavivirus RNA replication and screening for compounds inhibiting the viral RNA synthesis. Finally, we are analyzing cellular responses to expression of flavivirus encoded proteins and polyproteins.
Replication of SARS-CoV-2 and development of anti-coronavirus technologies
Since 2020 our research group has also started to work with SARS-CoV-2. We have constructed single plasmid based infectious cDNA clone for SARS-CoV-2 and use it to create tools for our studies of SARS-CoV-2 adaptations, resistance to antiviral compounds, functions of viral RNA structures and mechanism of virus-host interactions. We have generated >200 mutant versions of SARS-CoV-2 genome including version resistant to remdesivir (with mutation in nsP2) and number of life-attenuated vaccine candidates
-
Rihn, S.J., Merits, A., Bakshi, S., Turnbull, M.L., Wickenhagen, A., Alexander, A.J.T., Baillie, C., Brennan, B., Brown, F., Brunker, K., Bryden, S.R., Burness, K.A., Carmichael, S., Cole, S.J., Cowton. V.M., Davies, P., Davis, C., De Lorenzo, G., Donald, C.L., Dorward, M., Dunlop, J.I., Elliott, M., Fares, M., da Silva Filipe, A., Freitas, J.R., Furnon, W., Gestuveo, R.J., Geyer, A., Giesel, D., Goldfarb, D.M., Goodman, N., Gunson, R., Hastie, C.J., Herder, V., Hughes, J., Johnson, C., Johnson, N., Kohl, A., Kerr, K., Leech, H., Lello, L.S., Li, K., Lieber, G., Liu, X., Lingala, R., Loney, C., Mair, D., McElwee, M.J., McFarlane, S., Nichols, J., Nomikou, K., Orr, A., Orton, R.J., Palmarini, M., Parr, Y.A., Pinto, R.M., Raggett, S., Reid, E., Robertson, D.L., Royle, J., Cameron-Ruiz, N., Shepherd, J.G., Smollett, K., Stewart, D.G., Stewart, M., Sugrue, E., Szemiel, A.M., Taggart, A., Thomson, E.C., Tong, L., Torrie, L.S., Toth, R., Varjak, M., Wang, S., Wilkinson, S.G., Wyatt, P.G., Zusinaite, E., Alessi, D.R., Patel, A.H., Zaid, A., Wilson, S.J. & Mahalingam, S. (2021). A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biology, 19(2):e3001091. doi: 10.1371/journal.pbio.3001091.
-
Szemiel, A.M., Merits, A., Orton, R.J., MacLean, O.A., Pinto, R.M., Wickenhagen, A., Lieber, G., Turnbull, M.L., Wang, S., Furnon, W., Suarez, N., Mair. D., Filipe, A.S., Willett, B.J., Wilson, J.S., Patel, A.H., Thomson, E.C., Palmarini, M., Kohl, A., & Stewart, M.E. (2021). In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. , https://doi.org/10.1371/journal.ppat.1009929
-
Yang, S.L., DeFalco, L., Anderson, D., Zhang, Y., Aw, A., Lim, X.N., Tan, A.K.Y., Zhang, T., Chawla, T., Su, Y., Lezhava, A., Merits, A., Wang, L.-F., Huber, R. & Wan, Y. (2021). Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. ,
-
Ianevski, A., Yao, R., Zusinaite, E., Lello, L.S., Wang, S., Jo, E., Yang, J., Ravlo, E., Wan, W., Lysvand, H., Løseth, K., Oksenych, V., Tenson, T., Windisch, M.P., Poranen, M., Nieminen, A.I., Nordbø, S.A., Fenstad, M.H., Grødeland, G., Aukrust, P., Trøseid, M., Kantele, A., Lastauskienė, E., Vitkauskiene, A., Legrand, N., Merits, A., Bjørås, M. & Kainov, D.E. (2021) Synergistic interferon alpha-based combinations for treatment of SARS-CoV-2 and other viral infections. 2021;13(12):2489. doi: 10.3390/v13122489
-
Kubo, A.-L., Rausalu, K., Savest, N., Žusinaite, E., Vasiliev, G., Viirsalu, M., Plamus, T., Krumme, A., Merits, A. & Bondarenko, O. (2022). Antibacterial and antiviral effects of Ag, Cu and Zn metals, respective nanoparticles and filter materials thereof against coronavirus SARS-CoV-2 and Influenza A virus. 14, 2549. https://doi.org/10.3390/pharmaceutics14122549
We have efficient reverse genetics system for SARS-CoV-2 and variants of virus harboring marker gene. We do collaborate with research teams interested in genetic manipulation of SARS-CoV-2 genome to study an impact of mutations on the properties of the virus or possibilities of genetic attenuation the virus by introducing synonymous or missense mutations into coding regions of SARS-CoV-2 genome. We also participate in project aiming development of surfaces with virucidal properties that prevent surface transfer of SARS-CoV-2 and other viruses.
Ongoing and starting research projects/grants
-
Role of arbovirus replicase proteins in RNA replication, virus-host interactions and vector transmission (PRG1154, Estonian Research Council, 01/01/21-12/31/25), PI Andres Merits.
-
Antiviral Countermeasures Development Center (Antiviral Drug Discovery (AViDD) Centers, National Institute of Allergy and Infectious Diseases (NIAID), 05/01/22-04/30/25). PI
-
Synthetic reduced-vector-competence traits in Aedes aegypti (The Wellcome Trust Collaboration Award 226721/Z/22/Z. 04/01/23-03/31/28), PI, Luke Alphey (University of York, UK); Andres Merits, co-PI
-
Surface Transfer of Pathogens (European Commission. Proposal ID 101057961; 09/01/2022-08/31/2026); PI Artemis Stamboulis (University of Birmingham, UK), Andres Merits, Estonian co-PI